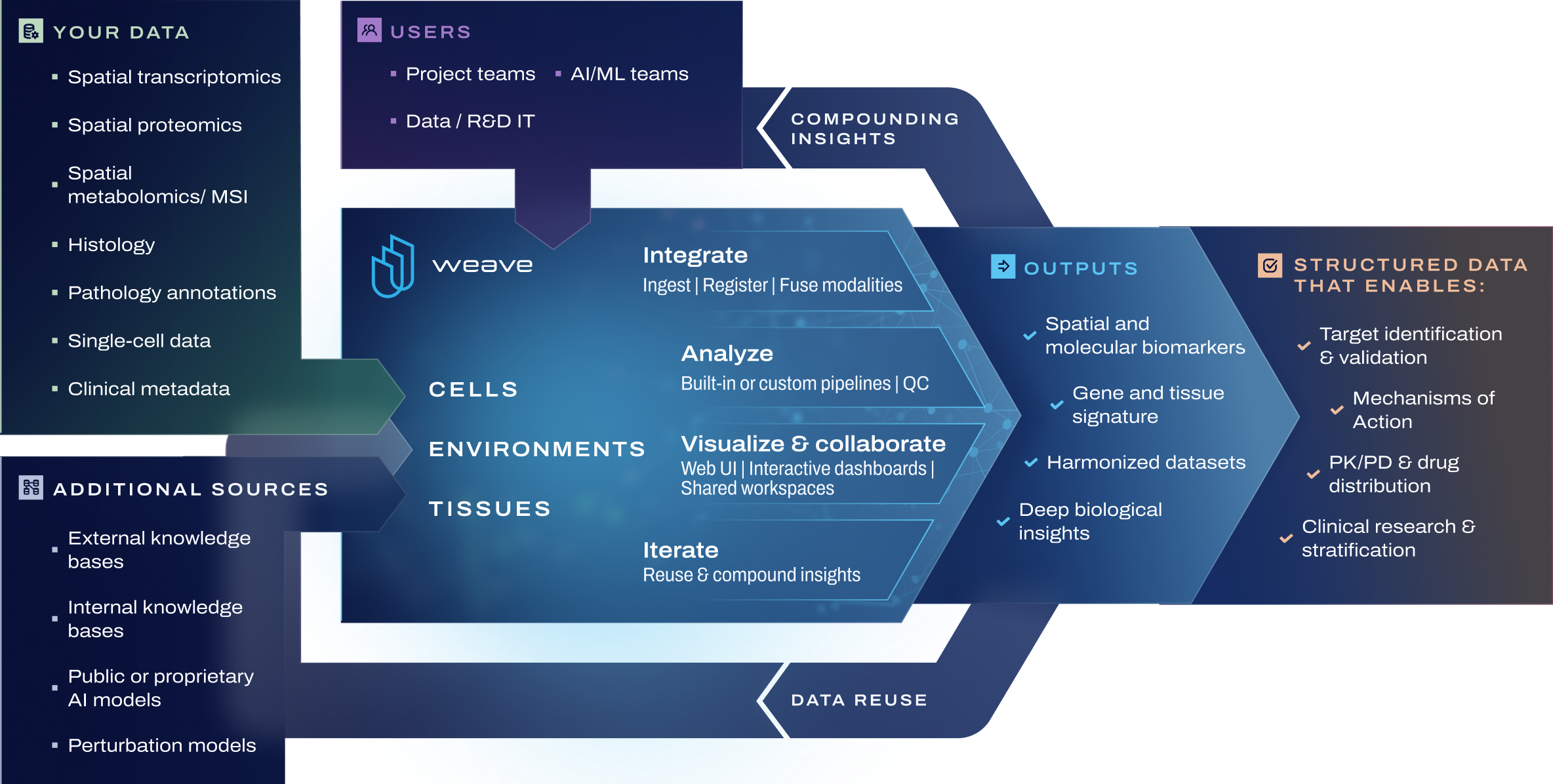
Your end-to-end platform for spatial and multimodal biological insight at enterprise scale
Weave unifies and structures spatial transcriptomics, proteomics, metabolomics, mass spectrometry imaging (MSI), single-cell data, histology, clinical metadata, and annotations into one governed workspace.
By preserving biological context across modalities, Weave enables analysis of cells, neighborhoods, and tissue architecture at scale.
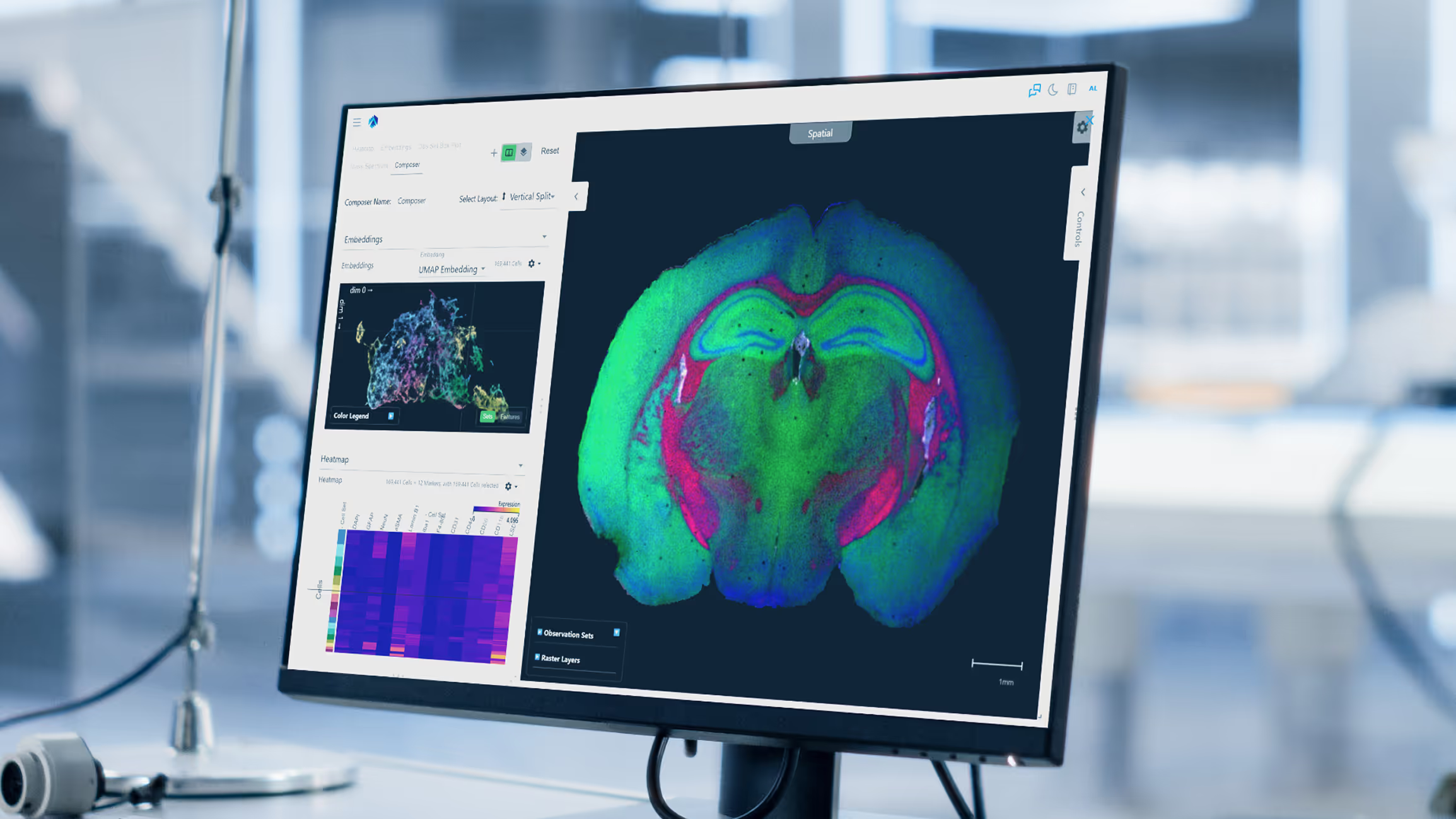
Weave combines spatial analysis with the flexibility to integrate your own computational pipelines. Interactively explore and share results, create and run routine analysis workflows, train AI models on structured data, and deploy outputs back into your wet-lab or computational workflows.
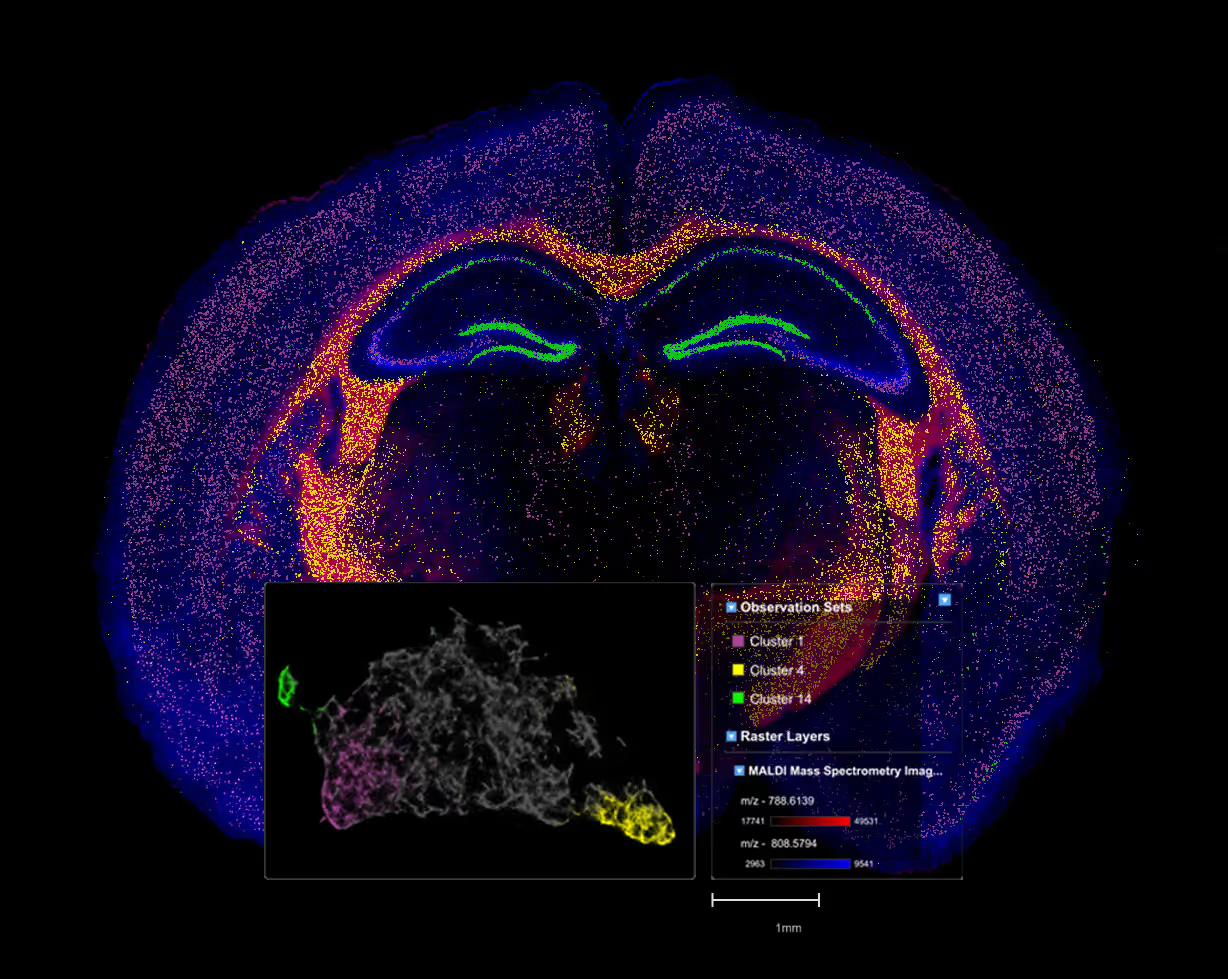
Explore multicellular tissue architecture using spatial omics
Spatial biology is multidimensional: multicellular, multimodal, and multi-disciplinary. Weave fuses data across these dimensions so scientists can focus on understanding biological mechanisms, relationships, and spatial context, instead of reconciling formats or pipelines.
Weave provides spatial context analysis, not just visualization
Identify, search, and annotate cellular neighborhoods and their functional behavior.
Study cell–cell interactions within relevant spatial neighborhoods.
Map ligand–receptor and pathway activity within true tissue context.
Detect treatment-driven phenotypic shifts earlier.
Connect morphological features with molecular changes.
Link biology to clinical metadata for stronger hypotheses.
The core concepts
Manage & integrate
Bring your spatial and molecular data into a coherent reference framework.
Analyze
Run built-in QC, segmentation, and neighborhood analysis, or integrate your own pipelines and models through our SDKs.
Visualize & collaborate
Interactively explore tissue structure, molecular signals, and cellular neighborhoods and share findings across teams, programs, and global sites.
Model & reuse
Train and deploy AI/ML models on governed datasets. Feed predictions and annotations back into Weave for traceable reuse and closed feedback loops.
Use Weave to build spatial data products
Weave provides consistent schemas, metadata standards, ontologies, and provenance tracking so data becomes structured, comparable, reusable, and model-ready across programs and stakeholders.
Structured data enables:
Reproducible analyses across sites, cohorts, and assay types
Governed collaboration with CROs, partners, and internal teams
Smooth integration with internal and external pipelines (LIMS, catalogs, DP tools)
Reliable AI/ML model development on consistent, domain-validated inputs
Closed-loop iteration between wet-lab and computation teams, reducing rework and turnaround time
Turn structured data into cumulative knowledge with Weave Insights
Your standardized datasets feed directly into Weave Insights, your spatiomolecular knowledge base that grows with every experiment.
Contextualize new data points with prior in-house or public knowledge
Validate findings without reprocessing old datasets
Assess mechanistic novelty across tissues, cohorts, or programs
Build a durable knowledge resource that compounds in value over time
This foundation allows you to build reliable, scalable workflows and generate results that hold up across studies and time, even as you add new cohorts, modalities, or research partners.
The Weave advantage in spatial biology
Deeper analysis
Combine spatial omics, histology, single-cell data, and metadata to expose mechanisms only visible in a spatial context. Analyze, search, and annotate cells as well as any combinations of these datasets.
Cross-team collaboration
Weave ensures your biology, pathology, and computational teams work in one environment with shared datasets and reproducible workflows.
AI-powered data reuse
You get governed, lineage-tracked datasets that scale from pilot studies to enterprise programs, AI model-ready for reuse across programs and pipelines. Easily scale from tens to hundreds of samples.
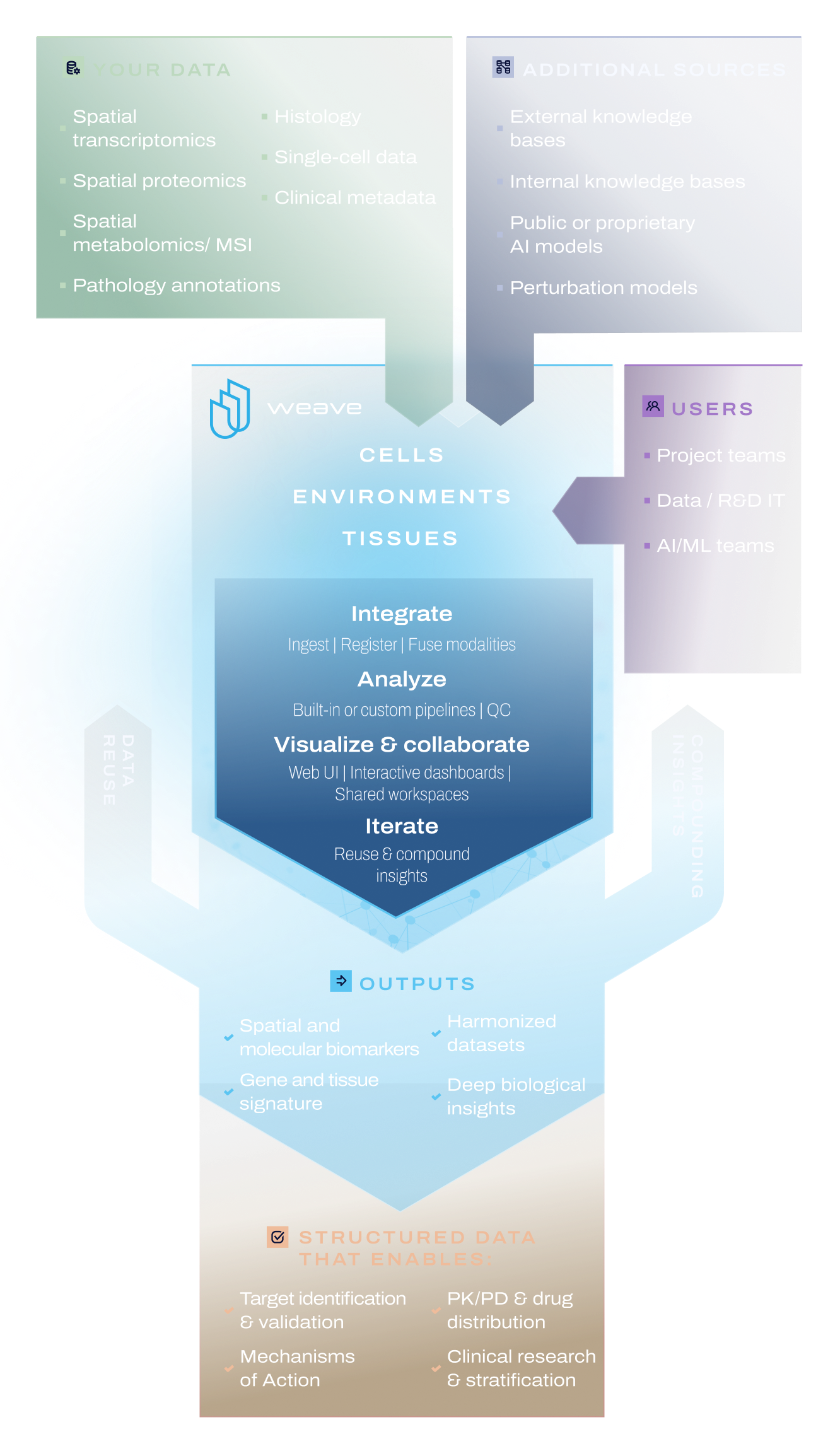
A governed platform for spatial biology, multimodal analysis, and AI workflows at enterprise-scale
.avif)
Unified data inputs
Ingest spatial transcriptomics, proteomics, metabolomics, MSI, histology, and single-cell data from any vendor for end-to-end processing and integrated analysis.
Core processing engine
Integrate multimodal spatial data while preserving spatial context, run QC, segmentation, and multicellular environment analytics, and turn raw readouts into reusable biological insight.
Enterprise data management
Govern your data with access controls, lineage, versioning and integrations into your catalogs, LIMS, and internal cloud environment.
Access & interfaces
Use the Weave web interface for visual analysis or connect through our SDK for custom pipelines and AI model development.
Use cases
End-to-end spatial multi-omics integration and analysis
Align histology with spatial molecular profiles to analyze biology in any dimension; powered by registration, patented multi-omics fusion, flexible cell annotation, QC, and reproducible workflows.
Scalable workflow implementation
Fit seamlessly into existing digital pathology, computational pipelines, and IT ecosystems without retooling.
Support for AI/ML workflows
Weave provides structured, domain-validated spatial datasets for AI/ML, enabling LLM-driven analysis, agentic RAG, and foundation model training. Its FAIR-aligned data foundation preserves biological context and maps insights into a reusable, enterprise-wide knowledge base.
Generate insights with Weave®
