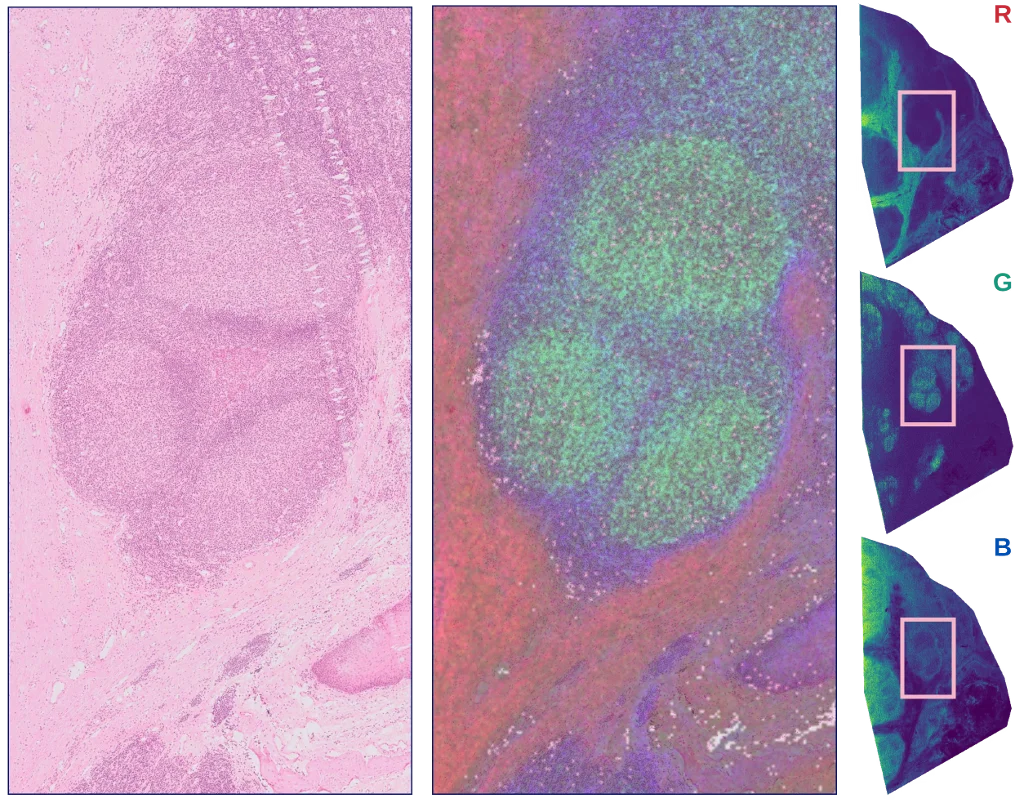
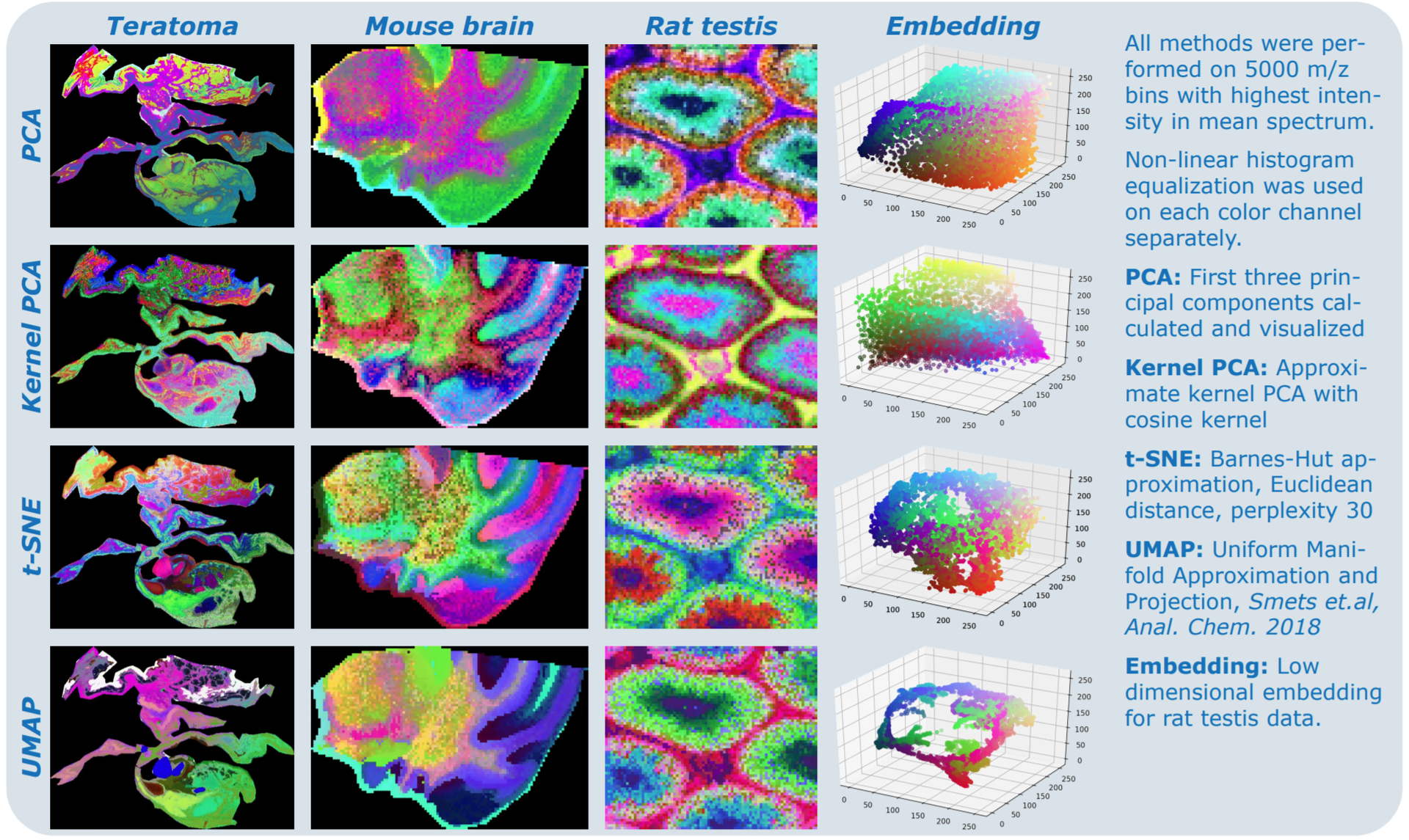
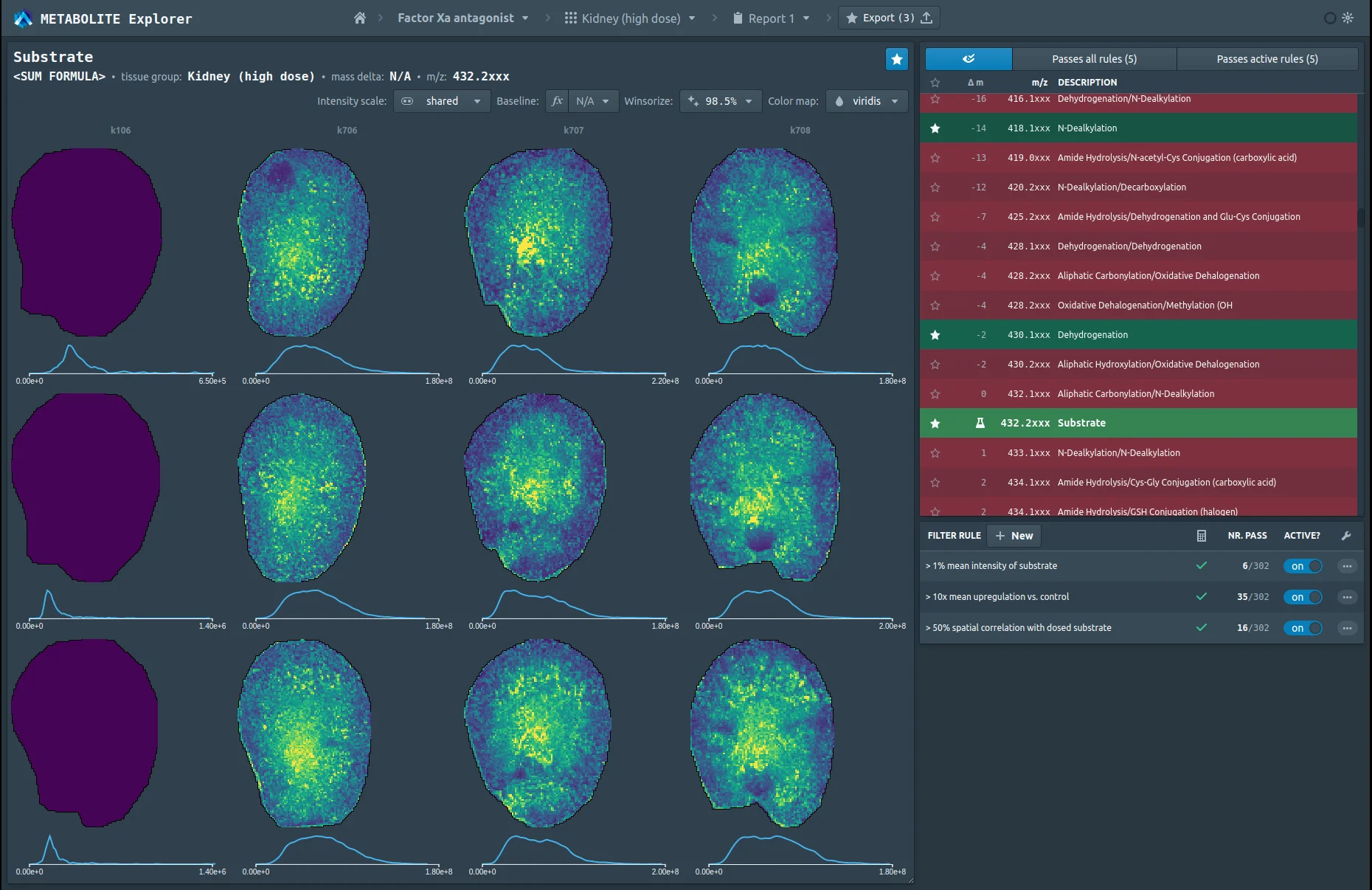
Explore peer-reviewed studies, technical notes, and posters that show how spatial and multimodal data can reveal mechanisms, guide decisions, and accelerate discovery.
Resources for advancing spatial insight
Learn from real project examples, methodological advances, and insights from our scientific team.
See how research teams use Weave to align modalities, analyze multicellular environments, validate mechanisms, and compare cohorts with reproducible workflows.
FAQs
Understanding Weave®, spatial biology, and multimodal analysis
Still have questions?
Weave supports multi-user, multi-team projects. With our shareable and compliant workspaces, different team members can collaborate in real-time on the same datasets in one governed workspace. Biologists, pathologists, computational biologists, and data leaders can interactively visualize and explore the same measurements and derived results, for both single and multi omics datasets. True democratization of spatial biology insights.
Yes. Browse our publication section above to explore peer-reviewed studies, application notes, posters, and collaborative projects demonstrating how Weave supports spatial and multimodal research across disease areas.
If your research involves tissue architecture, spatial biomarkers, spatial multi-omics integration, PK/PD readouts, or mechanistic interpretation, Weave provides a reproducible and scalable analytical framework. You can start with a single study and expand to multi-cohort programs.
Yes. Using the Weave SDK, our team can help you integrate your in-house pipelines, build reusable workflows, visualization modules, or reporting templates to reduce manual work and support high-throughput projects.
Yes. Aspect Analytics offers data analysis services from our expert team who have over a decade of experience in spatial bioinformatics and data analysis. Weave is entirely cloud-based, so you don’t need local processing power or specialized infrastructure to analyze spatial multi omics data. Taken together, our team can remotely support you with study-level data integration, cellular neighborhood analysis, workflow development and more, while your teams access the results and reusable workflows directly in the cloud. This collaboration keeps your projects moving even without in-house computing or engineering support.
Weave enables spatial PK/PD analysis, MoA exploration, responder vs. non-responder comparisons, and more. The platform produces deeper insights, and makes it easy to interpret and share across preclinical and translational groups. By providing a collaborative and secure environment, Weave turns massive, multi-modal spatial datasets into actionable insights to accelerate your drug discovery and development pipelines.
Weave provides the computational foundation to unify and analyze data across the multiple layers of spatial biology, histology, and metadata. It supports multimodal co-registration, comparative cohort analysis, model-ready outputs, and custom workflow development, bringing clarity to complex spatial data. Weave is also certified against the ISO 27001 and ISO 27018 standards for information security and handling of Personally Identifiable Information (PII).
Weave is used to explore cell-cell interactions, cellular neighborhoods & spatial niches, ligand–receptor communication, PK/PD and drug distribution, pathway activity, and tissue-level mechanisms that drive disease progression or therapeutic response. Weave enables finding spatial and/or molecular signatures indicative of disease status, treatment response, etc.
Weave standardizes quality control, spatial alignment and integration, and lineage tracking across sites, batches, and instruments. This creates reproducible, comparable datasets that support biomarker discovery, patient stratification, and translational analysis.
Yes. External transcriptomics, proteomics, MSI, or histology annotation files can be ingested and integrated. You can also load data that has been preprocessed using your in-house pipelines, or cell segmentation results from a pipeline you have optimized for your use-case. This lets you build complete, multi-layered datasets even when parts of the workflow happen elsewhere.
Yes. Aspect Analytics is vendor-neutral and works with all major spatial biology technologies. Supported platforms include Xenium, Visium, CosMx, COMET, PhenoCycler, IMC, assorted microscopy vendors, MALDI and DESI-MSI, and more. Core capabilities of Weave include analysis of large-scale projects using single or a limited number of spatial assays, or cross-platform harmonization across different modalities for comprehensive multi-omic characterization.
No. Biologists, pathologists, and computational scientists can all work within Weave. The Weave GUI supports visual analysis without coding, while the Weave SDK allows computational users to extend workflows or build custom analyses.
Weave software supports histology, spatial transcriptomics, spatial proteomics, multiplex imaging, mass spectrometry imaging (MSI), and complementary single-cell omics at scale. These layers can be aligned, harmonized, and compared within one governed analytical backbone.