
Aspect Analytics works at the forefront of spatial biology. Whether you’re running your first spatial study or operationalizing high-throughput, standardized analytical workflows for large-scale, multi-cohort programs, our scientists work directly with your biology, pathology, and computational teams.
Our services help you push the boundaries of spatial biology
Our services help you
Accelerate spatial and multimodal analysis
Build reproducible workflows inside Weave
Extend Weave with custom logic, pipelines, and visualizations
Train your team to operate Weave confidently across studies

Your extended
team of experts
Spatial biology creates more data than many teams can analyze or operationalize on their own. We help you turn your multimodal data into structured, reusable insight. We’ll handle whatever you need, from relieving the burden of routine repetitive analysis, to the most complex multimodal integration, exploratory algorithms, or high-end bioinformatics.
Whether you’re an early adopter or an innovator, we’ve got you covered.
The value we offer our customers:
Vendor-neutral expertise across all major spatial platforms
Deep experience with histology, spatial transcriptomics, proteomics, multiplex IF, MSI, and IMC
Extensive expertise with modality-specific complexity, non-linear relationships across layers, and multicellular environment analysis
Collaboration that spans biology, pathology, and computational science
Reusable, governed outputs instead of one-off reports
Transparent workflows your team can operate independently
Analytical and operational scalability for multi-omics programs and high-volume single-modality studies
Hands-on
Weave® training
We teach your pathologists, biologists, and computational scientists to run analyses, interpret results, and extend Weave workflows with confidence.
Depending on your background and needs,
you can access Weave on two levels:
Weave GUI
A visual interface for exploring spatial data, running analyses, and reviewing results without writing code.
Weave SDK
A Python toolkit for extending Weave with custom logic, advanced pipelines, and specialized analytical steps.
What this training enables:
Independent analysis across teams
Faster iteration without bottlenecks
Reuse of governed workflows across studies
Confident handover of analyses as your team scales its capabilities
And for those pushing the frontiers:
Development of custom analytical logic and modular pipelines for novel biological questions
Integration of exploratory algorithms or emerging AI methods into Weave’s governed workflows
Extension of multimodal analyses across layers, cohorts, and spatial scales
Confident use of the Weave SDK for high-end, custom-built components and workflows
We support your research in a wide range of areas
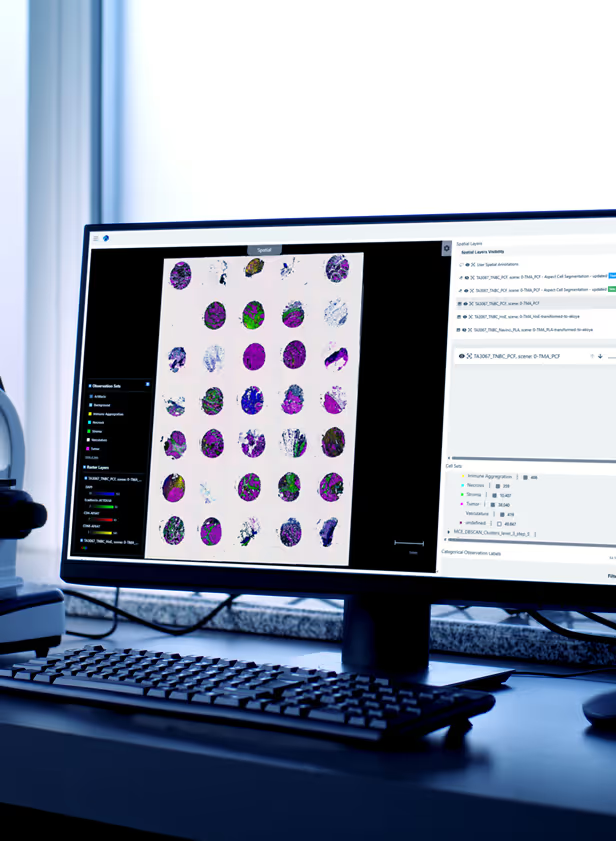
We’ll handle the full spectrum of integration work: aligning modalities, ingesting external data, applying QC and lineage tracking, and building governed assets your teams can reuse across studies. Align spatial and molecular layers while preserving biological context and addressing modality-specific constraints to support any scale of study, from multi-omics programs to large single-modality cohorts.
Alignment and harmonization of your histology, spatial transcriptomics, proteomics, metabolomics, MSI, and multiplex imaging data
Ingestion of external or pre-processed datasets, such as transcriptomics, proteomics, or MSI data, to supplement your results
Integrity checks, QC, and lineage to make sure reproducibility is maintained
Comparison of integrated data across your sites, batches, and instruments
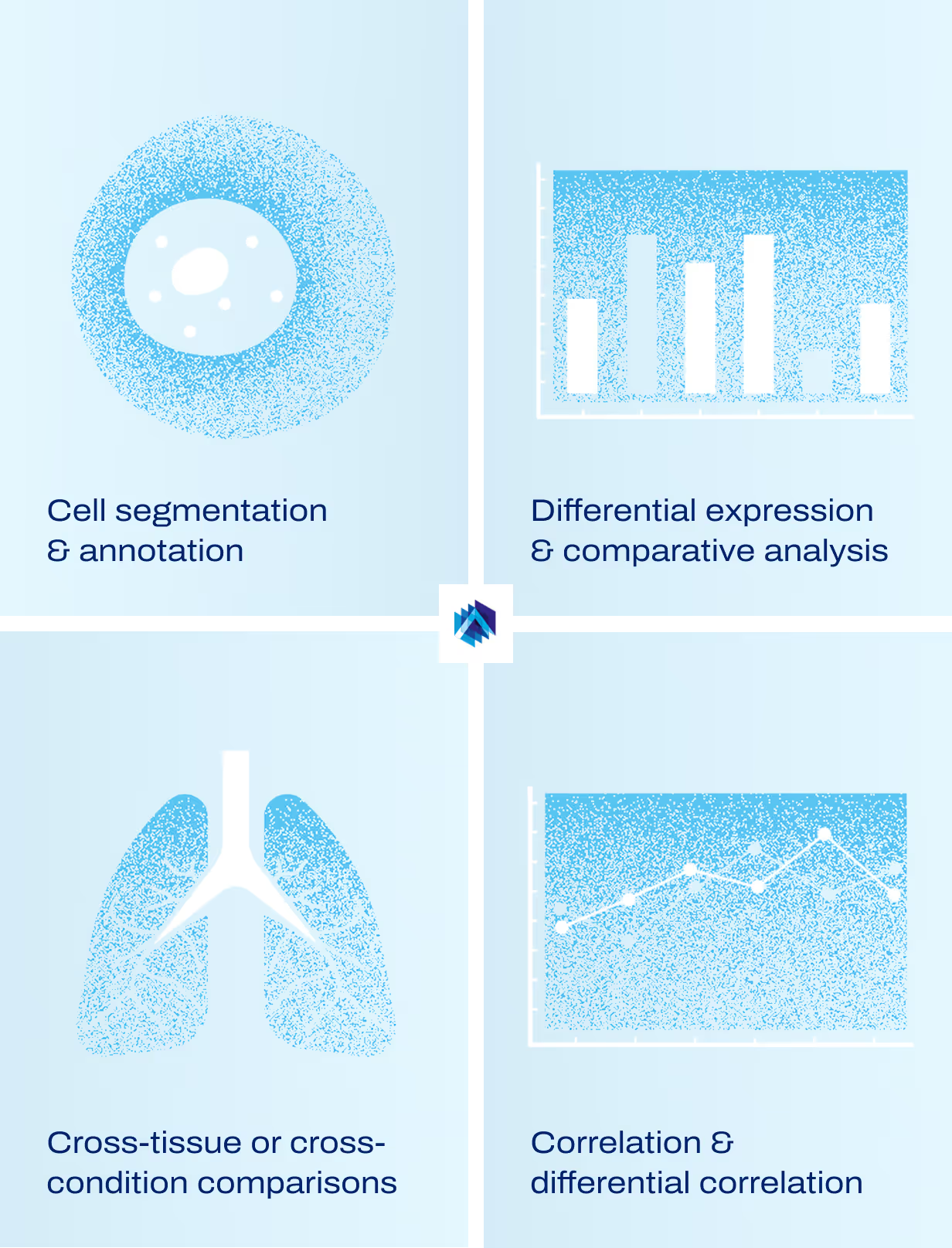
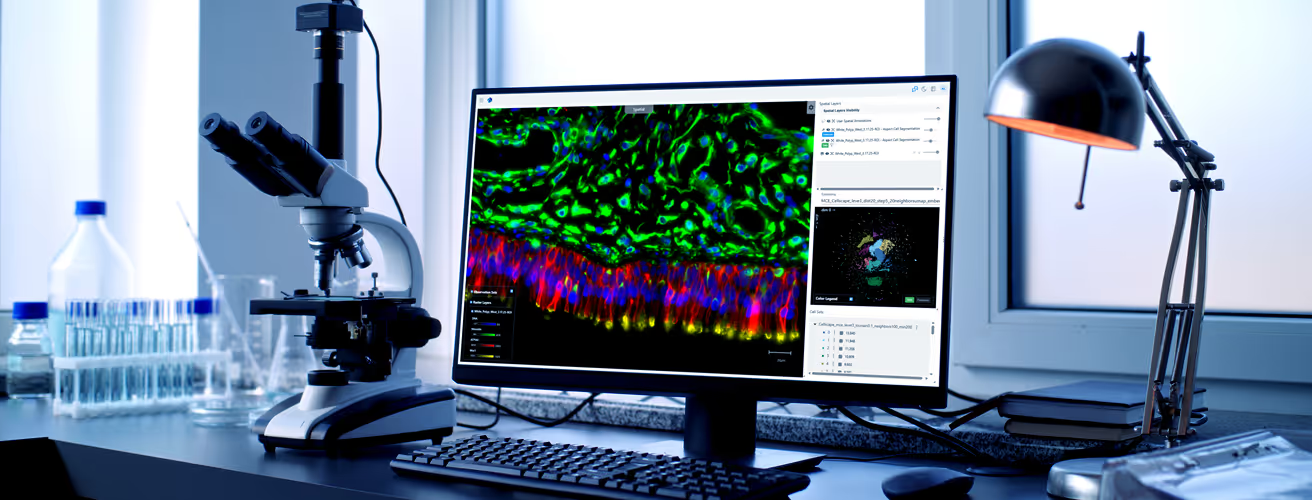
Core analytical capabilities
We’ll apply tailored analytical methods that match your biological question, whether you need standardized comparisons or advanced cross-modal interpretation. Our team scales with your needs from routine analytical workflows to complex, exploratory analysis across layers and cohorts. Clarify which molecular signals are meaningful, which patterns are noise, and where biological change is unfolding inside the tissue.
Identification, interpretation, and comparison of key cellular neighborhoods of interest
Cell-cell interactions and spatial dependencies to elucidate how cells influence one another in their local tissue context
Differential expression and comparative analysis of all available spatial layers across biological conditions
Correlation and differential correlation analysis to explore cross-omic relationships between your sample groups
Feature-level and region-level comparisons across tissues, conditions, or cohorts
Multicellular & cell-cell biology
Reveal how cells organize, communicate, and shape tissue-level phenotypes. We’ll help you move beyond marker-level analysis by defining and performing analytical methods that explore the microenvironments that shape disease progression and treatment response. We support everything from routine interaction analysis to advanced modeling of multicellular environments across modalities and scales.
Cellular neighborhood analysis to identify recurring spatial niches and reveal how the microenvironment influences disease progression
Cell-cell interactions and spatial dependencies to elucidate how cells influence one another in their local tissue context
Ligand–receptor communication analysis to reveal intercellular communication networks
Characterization of specific cell types in defined contexts, including localization, expression, and functional states
Quantification of cell positioning and migration relative to tissue structures and regions of interest
Translational & drug development analysis
Combine spatial context with molecular readouts to understand exposure, effect, and mechanism across preclinical and translational programs. Our capabilities range from routine PK/PD analyses to advanced multimodal co-registration linking drug localization with molecular and phenotypic shifts. Make informed preclinical and translational decisions with spatially resolved evidence.
PK/PD and drug distribution studies combining MSI-based drug localization with transcriptomic, proteomic, and metabolomic layers
Mechanism of action analysis linking pathway shifts to tissue architecture
Cross-cohort comparisons of treated vs. control samples
Identification of spatially resolved molecular signatures that can discriminate between patients (e.g., responders vs. non-responders) for precision medicine
Custom Weave development
Create workflows and components that match your exact scientific needs and automate recurring analyses, so your team can move faster without relying on ad hoc scripts or manual reprocessing. The resulting tailored Weave environment will operationalize your best practices and accelerate every program.
We’ll help you build:
Custom functionalities and analysis pipelines
SDK/API scripts for specialized calculations
Visualization modules and reporting templates
Scalable, governed workflows your team can run without ongoing external support
