Recap - AACR 2025
We attended the 2025 American Association for Cancer Research meeting in Chicago, with multiple presentations demonstrating how Weave supports researchers using spatial biology to understand cancer.
Access publication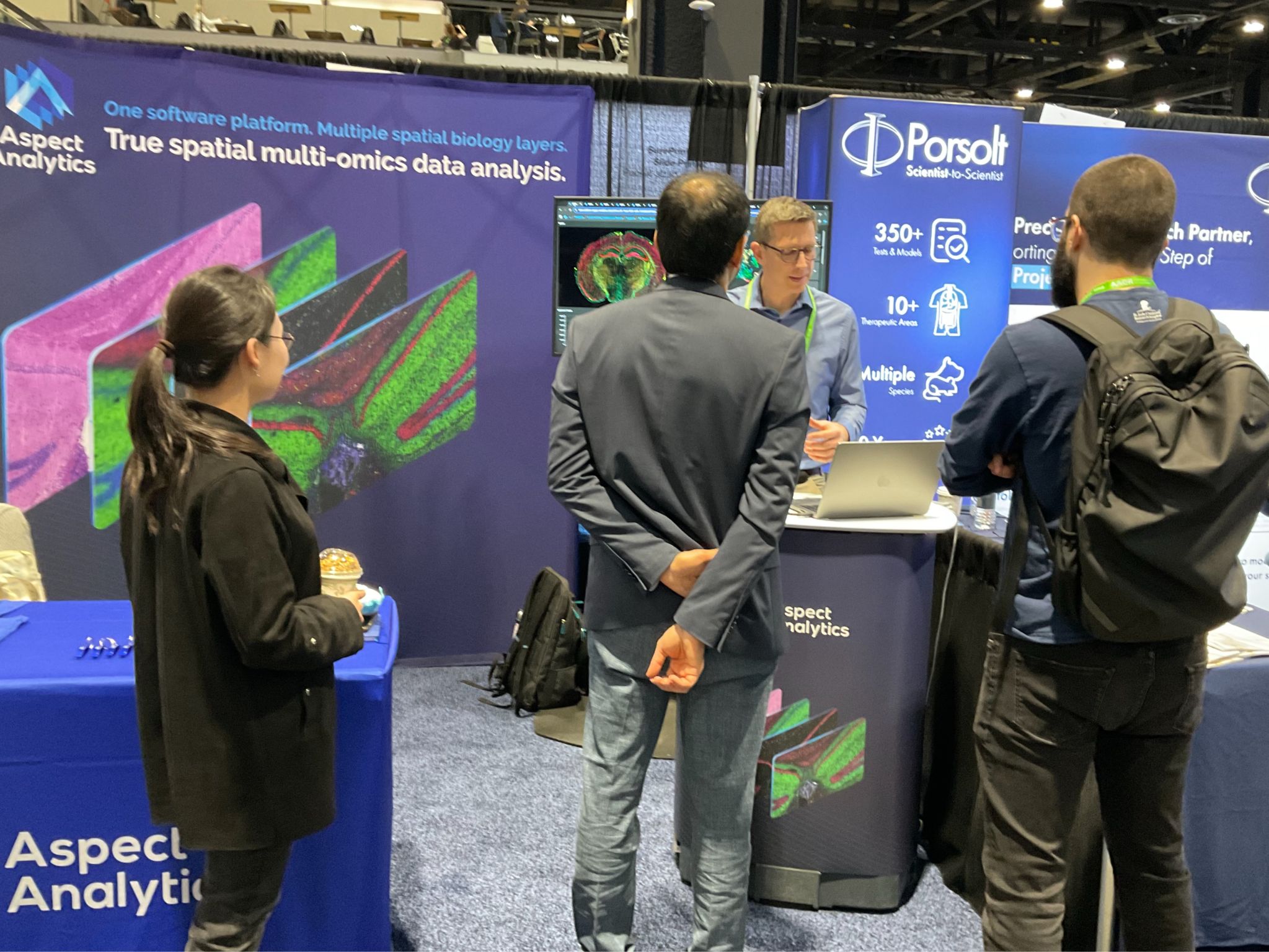
Aspect Analytics attended the AACR (American Association for Cancer Research) meeting in Chicago April 27-30, 2025. As cancer is characterized by significant changes to tissue architecture, it is no surprise that using spatial biology techniques to map the tumor microenvironment (TME) is of great interest in oncology research. The team presented how our Weave cloud platform enables integration of spatial multiomics data to help researchers understand the molecular and cellular mechanisms of development or resistance of disease and, ultimately, to match appropriate therapies to patients.
Aspect Analytics were co-authors on the following AACR posters:
“Integrating spatial metabolomics and spatial transcriptomics on the same cancer tissue sections to detect gene-metabolite correlations”

Trevor Godfrey (Baylor College of Medicine) presented poster abstract 150, where the location and abundance of hundreds of metabolites and mRNA were mapped from the same human cancer tissue sections. In this study, DESI mass spectrometry imaging for metabolites and Visium spatial transcriptomics were combined into one workflow. Data from these methods were co-registered and analyzed in Weave, enabling identification of thousands of spatial correlations between mRNA and metabolites.
This poster can be found here.
“In-situ Characterisation of Small Blue Round Cell Tumour Proteome and N-Glycome”

This collaboration with the Institute of Pathology, TU Munich, is a pilot study using a multimodal mass spectrometry imaging approach to distinguish between Small Round Blue Cell Tumours (SBRCT) subtypes. An umbrella term for tumors which are histologically similar but arise from different tissues, correct diagnosis of SBRCT is required for applying appropriate treatment. MALDI-MSI was used to spatially profile SBRCT samples, and different classification algorithms applied to determine if the tumor types could be distinguished via tryptic peptide, N-glycome or, after using Weave to integrate the datasets, a combined peptide-N-glycome profile. The findings demonstrated that the combined profile had the greatest ability to differentiate between SBRCT classes than via N-glycans or peptides individually.
To view this poster, please go here.
In addition, Colles Price from Takeda presented poster abstract 2418, “CellClique: Dissecting tumor microenvironments at the single cell level using generative AI and spatial transcriptomics” where Aspect Analytics provided data registration services, integrating a data analysis output from Path.AI with spatial transcriptomics. For AACR 2025 conference attendees, the e-poster can be found at: https://cattendee.abstractsonline.com/meeting/20273/Presentation/3248

Reach out if you have questions about the above studies, or want to discuss how we can support your own spatial multi-omics projects.
Images © 2025 Aspect Analytics. All rights reserved.