Quantitative MALDI Imaging of Aspirin Metabolites in Mouse Models of Triple-Negative Breast Cancer
This study refined quantitative MALDI imaging (QMALDI) to map aspirin metabolites, especially salicylic acid (SA). Precise spatial alignment, integration, and quantification of QMALDI, histology, and immunofluorescence images allowed accurate evaluation of the spatial distribution of SA in tissue regions, and helps with the further development of aspirin as an activatable MRI contrast agent.
Access publication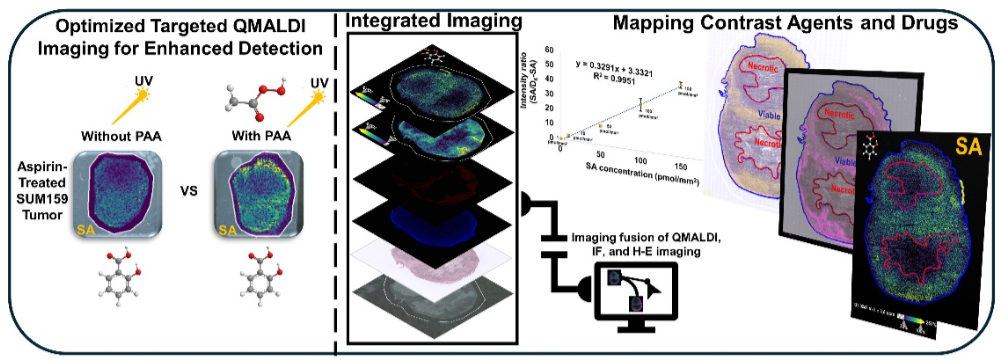
Aspirin and its active metabolites, such as salicylic acid (SA), are currently being developed as an activatable contrast agent for chemical exchange saturation transfer (CEST) magnetic resonance imaging (MRI). Researchers from The Johns Hopkins University School of Medicine refined a quantitative matrix-assisted laser desorption/ionization imaging method (QMALDI) to map aspirin metabolites in triple-negative breast cancer (TNBC) mouse models. The team developed a peracetic acid (PAA)-assisted QMALDI approach using a norharmane (nH) matrix for optimized sensitivity and uniformity. PAA was sprayed onto tissues to improve matrix consistency and reduce crystal size through hydrogen bonding with the matrix. Ultraviolet (UV) irradiation during imaging activated PAA, generating radicals that reduced background noise by breaking down matrix-related compounds.
Using Weave, integration of QMALDI data with histological and immunofluorescence imaging enabled precise spatial analysis of SA distribution. Results showed pronounced SA accumulation in the kidney medulla, liver, and viable tumor rims rich in blood vessels. This study demonstrates that the improved QMALDI imaging platform offers accurate, high-resolution visualization and quantification of drug metabolism and distribution in tissue. These findings highlight how integration of QMALDI, histology and immunofluorescence provides a comprehensive framework for studying drug delivery, metabolism, and tissue-specific localization of pharmacological compounds.
Please contact us if you are interested in learning more about this or have similar workflow needs.
Publication Details: Tae-Hun Hahm1,2, Dalton R. Brown1, Caitlin M. Tressler1,2, Thao Tran3, Alice Ly3, Arvind P.Pathak2,4,5, Michael T. McMahon2,6, Kristine Glunde1,2,5,7*
Quantitative MALDI Imaging of Aspirin Metabolites in Mouse Models of Triple-Negative Breast Cancer. Theranostics, 2025.
https://doi.org/10.7150/thno.116819
AFFILIATIONS
1 The Johns Hopkins Applied Imaging Mass Spectrometry Core and Service Center, Division of Cancer Imaging Research,
2 The Russell H. Morgan Department of Radiology and Radiological Science, The Johns Hopkins University School of Medicine, Baltimore, MD 21205, USA
3 Aspect Analytics NV, C-mine 12, 3600 Genk, Belgium
4 Department of Biomedical Engineering, The Johns Hopkins University School of Medicine, Baltimore, MD 21205, USA
5 The Sidney Kimmel Comprehensive Cancer Center, The Johns Hopkins University School of Medicine, Baltimore, MD 21205, USA
6 F.M. Kirby Research Center for Functional Brain Imaging, Kennedy Krieger Institute, Baltimore, MD 21205, USA
7 Department of Biological Chemistry, The Johns Hopkins University School of Medicine, Baltimore, MD 21205, USA