An integrated approach for analyzing spatially resolved multi-omics datasets from the same tissue section
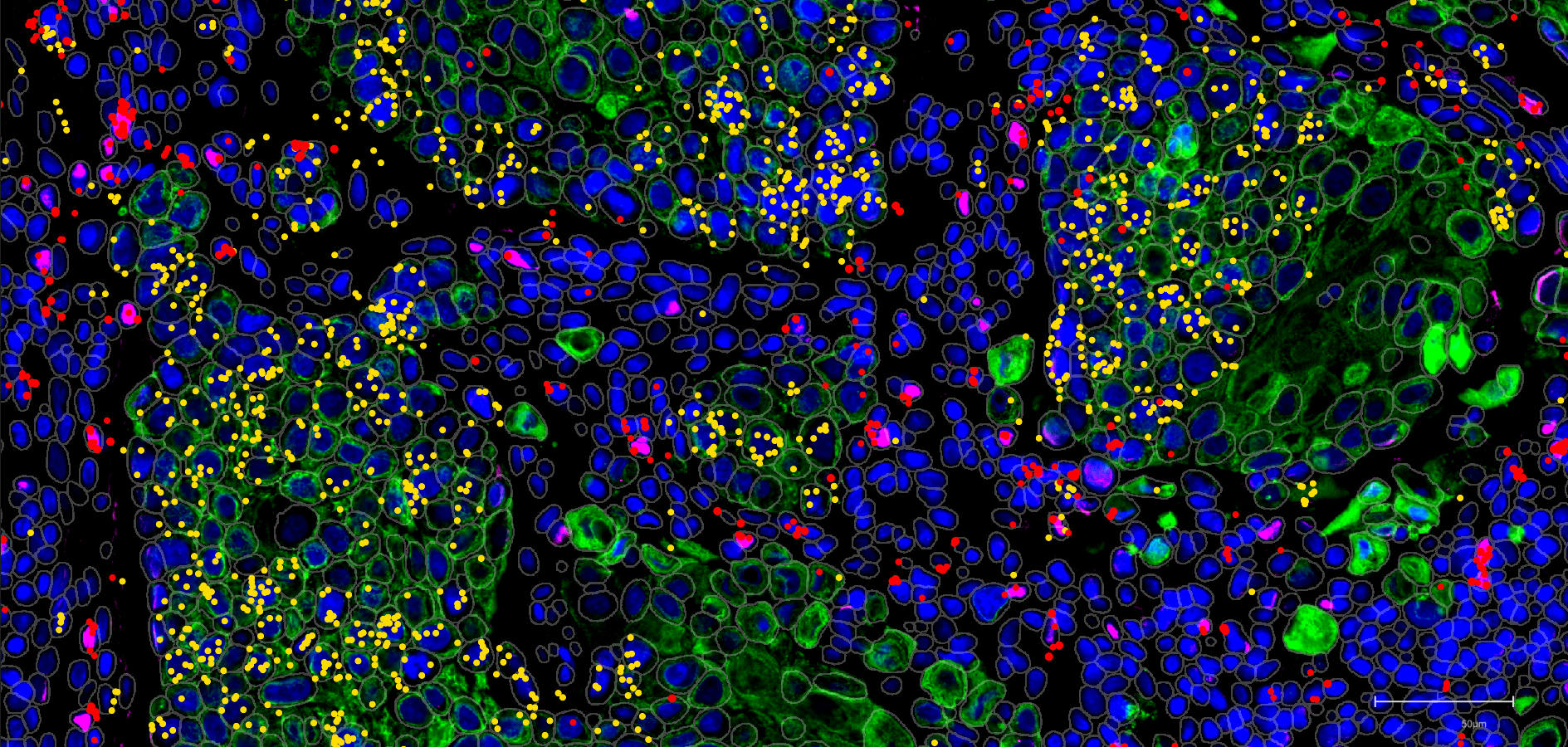
Recent advances in individual spatialomics technologies —such as spatial transcriptomics, proteomics, and metabolomics—have significantly enhanced our understanding of tissue biology. As these technologies evolve, combining multiple spatial readouts is becoming more common, though it presents computational and bioinformatics challenges. In this paper, we present a computational framework designed for integrating and analyzing same-section spatial transcriptomics and proteomics data acquired from different platforms, as demonstrated in our Weave software.
Using human lung cancer biopsy samples, tissue sections were sequentially analyzed using:
- Xenium (10X Genomics) for spatial transcriptomics (289-gene panel),
- COMET (Lunaphore) for spatial proteomics (40-antibody panel),
- and H&E staining for histopathology.
Weave software supported:
- non-rigid co-registration of the different datasets,
- integration of derived results from third party software (e.g. vendor cell segmentation, QuPath, HALO),
- interactive visualization of the integrated datasets at full resolution,
- communication of results between collaborators,
- and common downstream multimodal analysis pipelines.
This study demonstrates spatial integration of transcriptomic and proteomic data at the single-cell level, enabling evaluation of segmentation accuracy and quantification of RNA-protein correlations within individual cells.
Do not hesitate to contact us if you are interested in learning more about this or have similar workflow needs.
Publication Details: Thao Tran1, Felicia Wee2, Craig Ryan Joseph2, Wanqiu Zhang1, Jeffrey Chun Tatt Lim2, Zhen Wei Neo2, Li Yen Chong2, Francis Hong Xin Yap3, Nathan Heath Patternson1, Marc Claesen1, Alice Ly1* and Joe Yeong2,3,4*
An integrated approach for analyzing spatially resolved multi-omics datasets from the same tissue section. Front. Mol. Biosci., 2025, https://doi.org/10.3389/fmolb.2025.1614288
AFFILIATIONS
1 Aspect Analytics NV, Genk, Belgium,
2 Institute of Molecular Cell Biology (IMCB), Agency for Science, Technology and Research (A∗STAR), Singapore,
3 Department of Anatomical Pathology, Singapore General Hospital, Singapore
4 Department of Microbiology, Immunology and Serology Section, Singapore General Hospital, Singapore