

MSI applications part 1: glycans
In this blog post we introduce you to the wonderful world of glycans, an interesting analyte for which mass spectrometry imaging is proving to be particularly powerful.
Access publication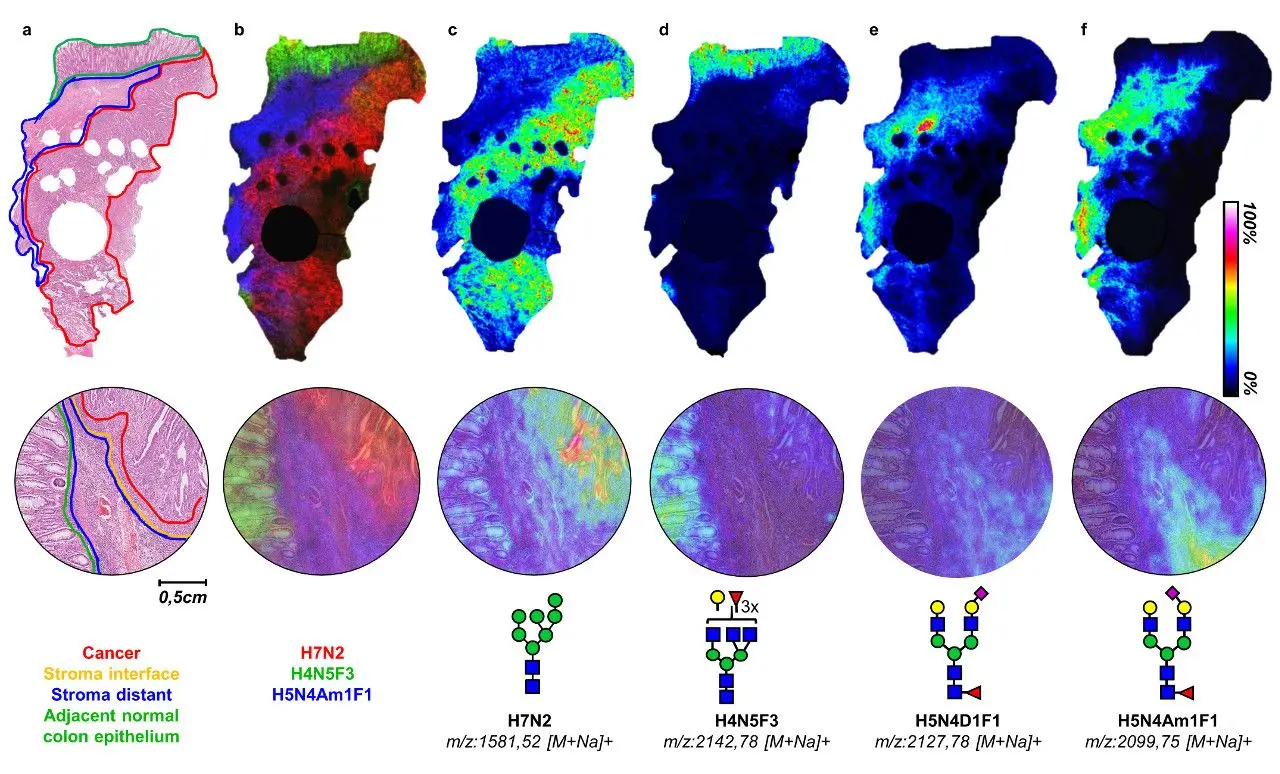

In this blog post we introduce you to the wonderful world of glycans, an interesting analyte for which mass spectrometry imaging is proving to be particularly powerful.
This post is part of our series titled "Introduction to mass spectrometry imaging", which contains the following entries:
- MSI hardware part 1: ionization techniques
- MSI hardware part 2: mass analyzers
- MSI applications part 1: glycans (current post)
- MSI applications part 2: peptides and proteins
- MSI applications part 3: lipids
- MSI applications part 4: drugs and small molecule metabolites
Table of contents
Introduction
Welcome back to our introduction to mass spectrometry imaging (MSI) series of blogs. Next topic in the series – applications! Some might say “Mass spectrometry imaging is just pretty pictures but doesn’t actually tell me much about the molecules compared to LC-MS/MS or established imaging techniques.” What these people fail to appreciate is that MSI adds an extra dimension to both of these (and other) techniques, which is perhaps best demonstrated by the analyte discussed in this blog – glycans.
What are glycans and why study them?
Glycans are polymers composed of monosaccharides linked by glycosidic bonds. These sugar-based chains act like sweet little blankets that coat the surfaces of cells and form a crucial part of each cell’s identity.
Glycosylation is the enzymatic attachment of glycans to other structures, such as lipids, and is a major post-translational modification of proteins. In eukaryotic cells, glycans are most commonly attached to proteins via a serine/threonine residue (O-linked glycosylation) or an asparagine residue (N-linked glycosylation). At the cellular level, glycans are involved in many basic cellular functions in mammalian cells such as protein folding, targeting, recognition, and cell adhesion, but also in immune surveillance and inflammatory responses.

Because glycan synthesis and glycosylation are highly regulated but not directly template-driven by the genome, changes in physiological and pathological states may be more readily reflected in glycans before gene mutations and protein expression. Alterations in glycosylation have been associated with different pathological conditions including gastrointestinal and liver disease,1 age-related illnesses,2 and the development and progression of different types of cancer.3
So why use MSI?
Mass spectrometry is the preferred method for the analysis of glycans as it allows characterization at different levels, from basic monosaccharide units to intact glycan-lipid complexes.4 However, the spatial distribution of molecules is lost when tissue is homogenized for LC-MS/MS analysis, which is an issue when using samples that contain a mixture of tissues and cells (e.g. cancer samples can contain tumor cells, scar tissue, inflammation, etc). While lectins, a type of carbohydrate-binding protein found in plants, are used for labelling glycan distribution in tissues, the mapping of specific glycans in histological and histopathological samples has been largely hampered by the lack of selective and sensitive commercial antibodies for their detection.
MALDI-MSI has emerged as a promising tool for investigating the spatial distribution of glycans in tissues due to the combination of aspects from both mass spectrometry and histology. The MALDI-MSI of N-glycans has been largely driven by the group of Richard Drake and colleagues from the Medical University of South Carolina, from the initial MALDI-MSI study using Peptide N-glycosidase F (PNGaseF) to enzymatically cleave N-glycans from frozen tissue sections,5 followed by the more successful application to formalin-fixed paraffin-embedded (FFPE) tissue sections and tissue microarrays (TMA),6 and subsequently to cultured cells and serum samples.7,8
Sample preparation for N-glycan analysis
A number of specific sample preparation steps are necessary to enable analyzing N-glycans with MSI. Without going into too much detail, the sample preparation for N-glycans generally follows the steps outlined below:
- Incubation and/or washing in solvents Depending on the type, the samples will either need to be delipidated (cultured cells, frozen tissue sections) or deparaffinized and rehydrated (FFPE samples).
- Heat-induced antigen retrieval This is similar to a step utilized in immunohistochemistry and is used to break formalin crosslinks between proteins in the tissues formed during tissue fixation and increase the signal of tissue glycans. For MALDI-MSI of glycans, slides are boiled in a low-pH buffer.
- PNGaseF enzyme application and digestion This enzyme cleaves N-glycans from proteins at asparagine residues and is applied to the sample in a thin, even layer using an automated spraying robot. Digestion by the enzyme is facilitated by incubating samples at 37°C in a humid chamber; the thin layer should prevent delocalization of the cleaved glycans from their native location/s.
- Matrix application alpha-cyano-4-hydroxycinnamic acid matrix (abbreviated as CHCA or HCCA) is typically used for N-glycan analysis. It is also deposited on the tissue in a fine layer to produce uniform sized crystals with a large enough wetness to extract the glycans, but not so large as to lead to delocalization.
- Measurement The majority of MSI glycan studies have used MALDI instruments (TOF and FTICR) in positive mode for detecting glycans with sodium adducts [M+Na], although there are some studies reporting the detection of glycans using DESI ionization.9,10
- Data analysis This can include assignment of glycans to specific peaks and statistical analysis of the overall data. N-glycan-measured samples can be histologically stained post-measurement which allows co-registration of the MSI data with histological annotations. Researchers can then link glycan distributions to specific tissue features, e.g. if different glycans are present in normal or diseased areas.
Like all techniques for the measurement of complex compounds, there are some glycans which cannot be easily detected by using the above listed steps. However, there are studies exploring ways to improve detection such as chemical derivatization steps to stabilize sialic acid sugars,11 and sequential enzyme digestion to target specific residue linkages or a different analyte or even to analyze different analytes from the same tissue section.12,13,14
Perhaps the most intriguing methodological developments for glycan measurement are MALDI-2 post-ionization to detect glycans in negative mode [M-H],15 and the use of ion mobility to separate isobaric glycans. Regular MSI experiments cannot currently distinguish glycans with multiple, different branches but the same molecular weight - they would appear as one peak - but ion mobility LC-MS/MS experiments have been successfully used to separate and identify such glycans.16
Impact
That’s interesting! But have there been any significant findings from MSI glycan studies? There sure have been! A recent review has examined glycan MSI in a clinical context including covering analysis on a variety of samples, but most of the work to date has examined glycans in a variety of cancer samples from FFPE tissues due to their presence in clinical laboratories, and hospital/university archives with patient data.17
Studies using MALDI-MSI of glycans found glycan signatures cannot only distinguish control tissues from cancer, but also potentially be used as markers for associated clinical events and outcomes. For example, a MALDI-MSI study of endometrial cancer using 20 patients without and 8 patients with lymph node metastases (LNM), glycans could discriminate between normal endometrial tissue, tumor, and tumors associated with lymph node metastases.18
A recent study of early-stage colorectal cancer using 16 patients not only found differences in the number of specific residues in glycans found in cancer vs. normal tissue, but there were also differences in the expression of glycans depending on whether they were found in tumor cells, tumor microenvironment in regions adjacent to or further away from the tumor, with particular glycan patterns in the adjacent microenvironment being associated with survival outcome (Figure 1).19 The results of this study are particularly striking as this kind of study is not currently possible via staining and the distribution information would have been lost using liquid mass spectrometric methods.

In a study of pancreatic ductal adenocarcinoma (PDAC), the authors conducted N-glycan MALDI-MSI analysis of 53 patients in a TMA with serial sections analyzed for two different biomarkers: CA19-9, the sole FDA-approved PDAC prognostic marker, and sTRA, a novel biomarker that appears on a non-identical subset of PDAC tumors to CA19-9.20 While the findings once again indicated that N-glycan content is different between healthy and diseased tissues, the CA19-9 and sTRA categorized tumor samples had different N-glycan content from each other. The extensive MALDI-MSI glycan data also allowed the authors to identify potential new biomarkers to discriminate PDAC from normal tissue, which were confirmed with lectin immunohistochemistry, thus showing the benefit of their combined MALDI-MSI and lectin-immunohistochemistry approach.
Conclusion
In this post, we introduced the glycans: what they are, why they’re important, how mass spectrometry imaging of glycans is achieved, and gave examples of how different groups have used MALDI-MSI of glycans to conduct clinically relevant studies. If this has piqued your interest for investigating glycans with mass spectrometry imaging, feel free to contact Aspect Analytics for data analysis options.

We encountered Glycandalf for the first time on reddit.
CONTACT US
References:
- Verhelst, X. et al. Protein Glycosylation as a Diagnostic and Prognostic Marker of Chronic Inflammatory Gastrointestinal and Liver Diseases. Gastroenterology 158, 95-110, doi:10.1053/j.gastro.2019.08.060 (2020).
- Paton, B., Suarez, M., Herrero, P. & Canela, N. Glycosylation Biomarkers Associated with Age-Related Diseases and Current Methods for Glycan Analysis. Int J Mol Sci 22, doi:10.3390/ijms22115788 (2021).
- Peixoto, A., Relvas-Santos, M., Azevedo, R., Santos, L. L. & Ferreira, J. A. Protein Glycosylation and Tumor Microenvironment Alterations Driving Cancer Hallmarks. Front Oncol 9, 380, doi:10.3389/fonc.2019.00380 (2019).
- Mass Spectrometry of Glycoproteins. (Humana, New York, NY, 2021).
- Powers, T. W. et al. Matrix assisted laser desorption ionization imaging mass spectrometry workflow for spatial profiling analysis of N-linked glycan expression in tissues. Anal Chem 85, 9799-9806, doi:10.1021/ac402108x (2013).
- Powers, T. W. et al. MALDI imaging mass spectrometry profiling of N-glycans in formalin-fixed paraffin embedded clinical tissue blocks and tissue microarrays. PLoS One 9, e106255, doi:10.1371/journal.pone.0106255 (2014).
- Angel, P. M. et al. A Rapid Array-Based Approach to N-Glycan Profiling of Cultured Cells. J Proteome Res 18, 3630-3639, doi:10.1021/acs.jproteome.9b00303 (2019).
- Blaschke, C. R. K., Black, A. P., Mehta, A. S., Angel, P. M. & Drake, R. R. Rapid N-Glycan Profiling of Serum and Plasma by a Novel Slide-Based Imaging Mass Spectrometry Workflow. J Am Soc Mass Spectrom 31, 2511-2520, doi:10.1021/jasms.0c00213 (2020).
- Pace, C. L. & Muddiman, D. C. Direct Analysis of Native N-Linked Glycans by IR-MALDESI. J Am Soc Mass Spectrom, doi:10.1021/jasms.0c00176 (2020).
- Skraskova, K. et al. Enhanced capabilities for imaging gangliosides in murine brain with matrix-assisted laser desorption/ionization and desorption electrospray ionization mass spectrometry coupled to ion mobility separation. Methods 104, 69-78, doi:10.1016/j.ymeth.2016.02.014 (2016).
- Holst, S. et al. Linkage-Specific in Situ Sialic Acid Derivatization for N-Glycan Mass Spectrometry Imaging of Formalin-Fixed Paraffin-Embedded Tissues. Anal Chem 88, 5904-5913, doi:10.1021/acs.analchem.6b00819 (2016).
- Clift, C. L., Drake, R. R., Mehta, A. & Angel, P. M. Multiplexed imaging mass spectrometry of the extracellular matrix using serial enzyme digests from formalin-fixed paraffin-embedded tissue sections. Anal Bioanal Chem 413, 2709-2719, doi:10.1007/s00216-020-03047-z (2021).
- Heijs, B. et al. Multimodal Mass Spectrometry Imaging of N-Glycans and Proteins from the Same Tissue Section. Anal Chem 88, 7745-7753, doi:10.1021/acs.analchem.6b01739 (2016).
- West, C. A., Liang, H., Drake, R. R. & Mehta, A. S. New Enzymatic Approach to Distinguish Fucosylation Isomers of N-Linked Glycans in Tissues Using MALDI Imaging Mass Spectrometry. J Proteome Res 19, 2989-2996, doi:10.1021/acs.jproteome.0c00024 (2020).
- Heijs, B., Potthoff, A., Soltwisch, J. & Dreisewerd, K. MALDI-2 for the Enhanced Analysis of N-Linked Glycans by Mass Spectrometry Imaging. Anal Chem 92, 13904-13911, doi:10.1021/acs.analchem.0c02732 (2020).
- Gao, Z. et al. Distinguishment of Glycan Isomers by Trapped Ion Mobility Spectrometry. Anal Chem, doi:10.1021/acs.analchem.1c01461 (2021).
- Blaschke, C. R. K. et al. Glycan Imaging Mass Spectrometry: Progress in Developing Clinical Diagnostic Assays for Tissues, Biofluids, and Cells. Clin Lab Med 41, 247-266, doi:10.1016/j.cll.2021.03.005 (2021).
- Mittal, P. et al. Altered N-linked glycosylation in endometrial cancer. Anal Bioanal Chem 413, 2721-2733, doi:10.1007/s00216-020-03039-z (2021).
- Boyaval, F. et al. N-glycomic signature of stage II colorectal cancer and its association with the tumor microenvironment. Mol Cell Proteomics, 100057, doi:10.1074/mcp.RA120.002215 (2021).
- McDowell, C. T. et al. Imaging Mass Spectrometry and Lectin Analysis of N-Linked Glycans in Carbohydrate Antigen-Defined Pancreatic Cancer Tissues. Mol Cell Proteomics 20, 100012, doi:10.1074/mcp.RA120.002256 (2020).
